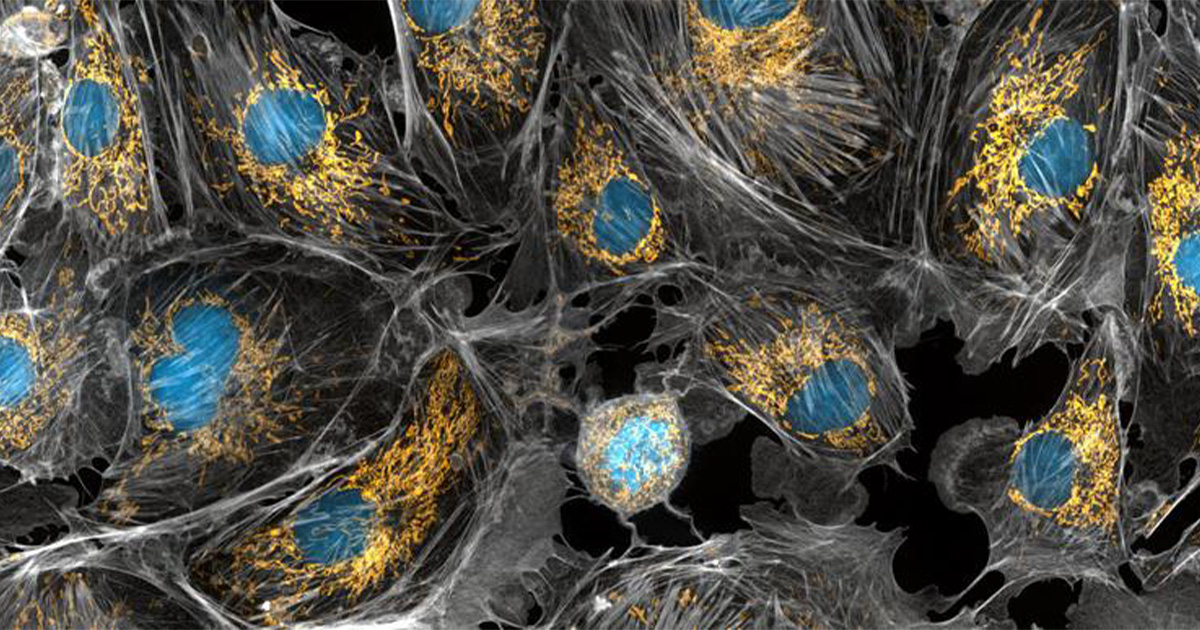
Before the first mitochondria were encapsulated by the earliest eukaryotic cells around 1.6 to 1.8 billion years ago, these aptly nicknamed "powerhouses of the cell" existed as free-living bacteria.
Now, a Science Advances study suggests the closest living relatives to mitochondria may belong to a genus of marine bacteria called Iodidimonas. This finding also means that the genus could be the closest ancestral link to mitochondria's forebearers, or protomitochondria.
Further analyses of Iodidimonas species could add insights as to what protomitochondria's ancient genome might have looked like. In turn, this could reveal clues about the evolutionary origins of eukaryotes, which represent most plants, animals and fungi.
"The most recent common ancestor from which all eukaryotes descended is termed the last eukaryotic common ancestor or LECA. All eukaryotes have mitochondria. So, we infer that the LECA also had mitochondria," explained Parth Raval, an evolutionary biologist at the University of Düsseldorf and first author of a related Science Advances Focus contextualizing the new discovery. "Mitochondria play key roles in eukaryotic cell biological processes, such as the cell cycle and cell death, across all eukaryotes. Hence, we suggest that it played a key role in shaping the LECA itself."
These results also challenge past investigations, which had championed a different genus called Rickettsia as protomitochondria's relatives.
"I was never convinced that Rickettsia and its relatives could be the likely ancestors of mitochondria," said Mauro Degli Esposti, a scientist at the Universidad Nacional Autonoma de Mexico (UNAM) and the corresponding author of the paper.
Origins of the Organelle
Around 1.8 to 1.6 billion years ago, mitochondria took up permanent residence in unicellular organisms called archaeons. This relationship yielded endless metabolic and structural benefits. Yet, it's hard to pin down the genetic instructions protomitochondria brought with them during this endosymbiotic event.
"As much as we would like to know what the mitochondrial ancestor looked like, it died two billion years ago," said Raval. "This leaves us with indirect approaches where we compare current mitochondrial features with current proteobacterial features to find a best match."
In the late 1990s, work tying the Rickettsia genus to protomitochondria had just been published. Rickettsia bacteria are intracellular anerobic parasites. The most famous species is tick-borne Rickettsia, which causes Rocky Mountain spotted fever. Because of the genus's intracellular and anerobic features, researchers thought it might contain vestigial traces of protomitochondria.
"In bacteria, we cannot have such fossils representing intermediate states," said Esposti. "However, we can look at genomic features that persist as vestigial remnants of biological systems that are either not used anymore or have been fragmented in today's genomes."
One of those vestigial remnants would be the ability to biosynthesize cardiolipin. This lipid coats the mitochondrial membrane and cannot be produced by unicellular eukaryotes. Genes to metabolize this lipid were irrefutably brought by protomitochondria into eukaryotic cell hosts. Intuitively, a history of those genes should then be detectable in the DNA of any possible mitochondrial relatives.
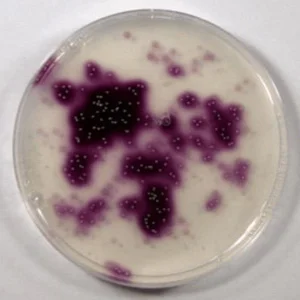 Iodidimonas strains isolated from natural gas brine water. (Image credit: NIH)
Iodidimonas strains isolated from natural gas brine water. (Image credit: NIH)
Esposti soon realized that Rickettsia species lacked the ability to biosynthesize cardiolipin as well as various other important metabolic traits. However, he struggled to find suitable alternate candidates within alphaproteobacteria. This widespread taxonomic class, which houses Rickettsia , has long been suspected to contain relatives of protomitochondria.
But the alphaproteobacterial class is extremely diverse and continues to be studied. In fact, Iodidimonas and the Iodidimonadales order were just discovered in 2016.
"[The Iodidimonas genus only] could be identified as a likely protomitochondrion candidate after sufficient information on its genome became available," said Otto Geiger, a specialist in ecological genomics at UNAM and the first author of the article.
As more taxonomic categories of alphaproteobacteria became known, Geiger and the group expanded their hunt for vestigial remains of protomitochondria.
Tracing Genetic Clues Across Time
For their first step, the researchers created a checklist of protomitochondria-specific metabolic traits. As expected, that criteria included the biosynthesis of cardiolipin. It also listed the metabolization of ceramide-based lipids, or sphingolipids. This skill is widespread in eukaryotes but rarely performed by bacteria.
"We also would expect that protomitochondria were able to form ceramide and maybe even complex sphingolipids," Geiger said. This expectation is, in part, because ceramides are crucial in helping form mitochondria's cristae, or their folded inner membranes.
Through genomic analyses, the team found that enzymes for ceramide biosynthesis in eukaryotes came from a bacterial ancestor within the alphaproteobacterial subclass called Caulobacteridae—an entirely different subclass from Rickettsidae, in which the previously championed Rickettsia genus belongs.
"Previous evolutionary speculations did not consider this likely possibility," said Geiger.
Beyond cardiolipin and sphingolipid biosynthesis, the scientists also screened for vestigial genomic units, including one called the COX operon. It's thought that alphaproteobacteria gained the COX operon via lateral gene transfer from a separate bacteria class. The operon let alphaproteobacteria survive better in in the increasingly oxygenated Proterozoic oceans. Today, gene clusters for the operon remain in living bacteria.
Detecting COX operons in Iodidimonas came with another unexpected result. Linked to the operon were genomic units directly connecting a mitochondrial structure called complex III to its long-suspected bacterial counterpart, the bc1 complex. Identifying the bc1 complex provided further proof that protomitochondria belonged in the Caulobacteridae subclass, not the Rickettsidae subclass.
Alongside this evidence, the group found that the Iodidimonadales order held a trove of genes that tied mitochondria to ancestral bacteria. They showed that the Iodidimonas genus order could conduct sphingolipid and cardiolipin biosynthesis. All signs pointed to the Iodidimonas genus being the living descendants of protomitochondria.
These takeaways also enhance the story of unicellular eukaryote evolution. Eukaryotic cells use ceramide and cardiolipin biosynthesis to conduct mitochondria signaling and to induce a form of mitochondria-specific autophagy. This cellular process allows cells to recycle their defective and unnecessary parts. Like autophagy, mitophagy is the self-digestion and recycling of mitochondria.
"[Mitophagy] was modified, or co-opted, to digest not only the internal particles, but engulfed prokaryotic food particles as well, and such a feeding lifestyle called phagotrophy became feasible," said Raval, assessing the study's implications. Engulfing protomitochondria, he explained, gave host cells the ability to ingest other nutritious particles and evolve cellular complexity to expedite that digestive process.
Protomitochondria essentially gave their early LECA hosts a mitophagy toolkit that eventually aided the transformation into all that the Eukarya domain encompasses today.
"From LECA, all of the rather diverse eukaryotic branches descended," Raval summarized.
From the Lab to the Hot Springs
Moving forward, Esposti, Geiger and their colleagues plan to start sampling species within the Iodidimonas genus to narrow their search for the closest living descendants of protomitochondria and today's mitochondria relatives. Esposti is working with Alejandro Sanchez-Flores, a co-author of the study and metagenomics specialist at UNAM, to create molecular probes that could better identify Iodidimonas members in geothermal springs.
"The most promising protomitochondria relatives of the Iodidimonadales lineage have been found in Japanese springs of geothermal origin, so far," said Esposti. "Mexico is rich in springs of this kind and the microbial community of some of these springs has been explored already, albeit superficially."
Geiger, meanwhile, plans to compare the enzymes that biosynthesize cardiolipin and ceramides in the Iodidimonas bacteria to those found in unicellular eukaryotes. He said doing so will help the group "to learn more on the specific evolution of these pathways upon undergoing endosymbiotic transformation," examining how protomitochondria's metabolic skillset was beneficial as it became embedded in host cells.
Of course, this is not the end of the intrigue that surrounds protomitochondria, they acknowledged.
"Most likely, sophisticated phylogenetic studies will come out to challenge the proposal of our study," Esposti said. "And so, the debate on which bacteria originated mitochondria is likely to go on until, one day, clear experimental evidence will emerge."